Chapter 5 Evaluate T cell modulatory BAs in patients with GVHD vs controls (figure 4)
5.1 3oxoLCA
filtered_combined_table %>%
left_join(cohort_BAS) %>%
filter(later=="Y") %>%
filter(ursodiol=="Y") %>%
ggplot(aes(y=log10(dehydrolithocholic_acid+25000), x=GI_GVHD, color=GI_GVHD))+
geom_boxplot(width=0.2, lwd=0.8, outlier.shape = NA) +
geom_jitter(width=0.3, alpha=0.3, size=2.5)+
ylab("log10(3oxoLCA)")+
xlab("")+
theme_classic()+
stat_compare_means(comparisons=list(c("Y", "N")),
method="wilcox.test",
correct=FALSE)+
scale_color_manual(values=c("dodgerblue4", "red3"))+
theme(legend.position="none")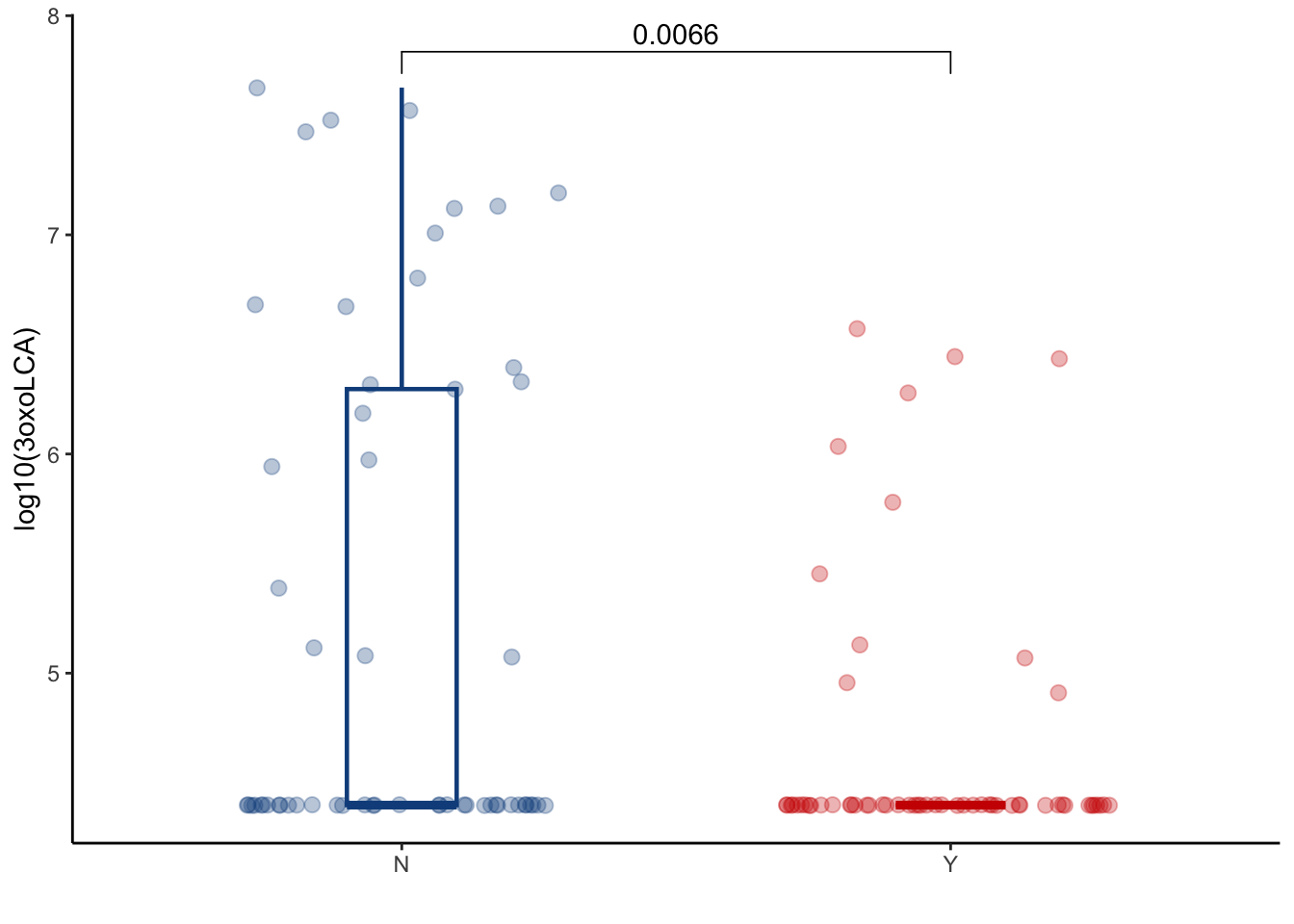
5.2 isoLCA
filtered_combined_table %>%
left_join(cohort_BAS) %>%
filter(later=="Y") %>%
filter(ursodiol=="Y") %>%
ggplot(aes(y=log10(isolithocholic_acid+25000), x=GI_GVHD, color=GI_GVHD))+
geom_boxplot(width=0.2, lwd=0.8, outlier.shape = NA) +
geom_jitter(width=0.3, alpha=0.3, size=2.5)+
ylab("log10(isoLCA)")+
xlab("")+
theme_classic()+
stat_compare_means(comparisons=list(c("Y", "N")),
method="wilcox.test",
correct=FALSE)+
scale_color_manual(values=c("dodgerblue4", "red3"))+
theme(legend.position="none")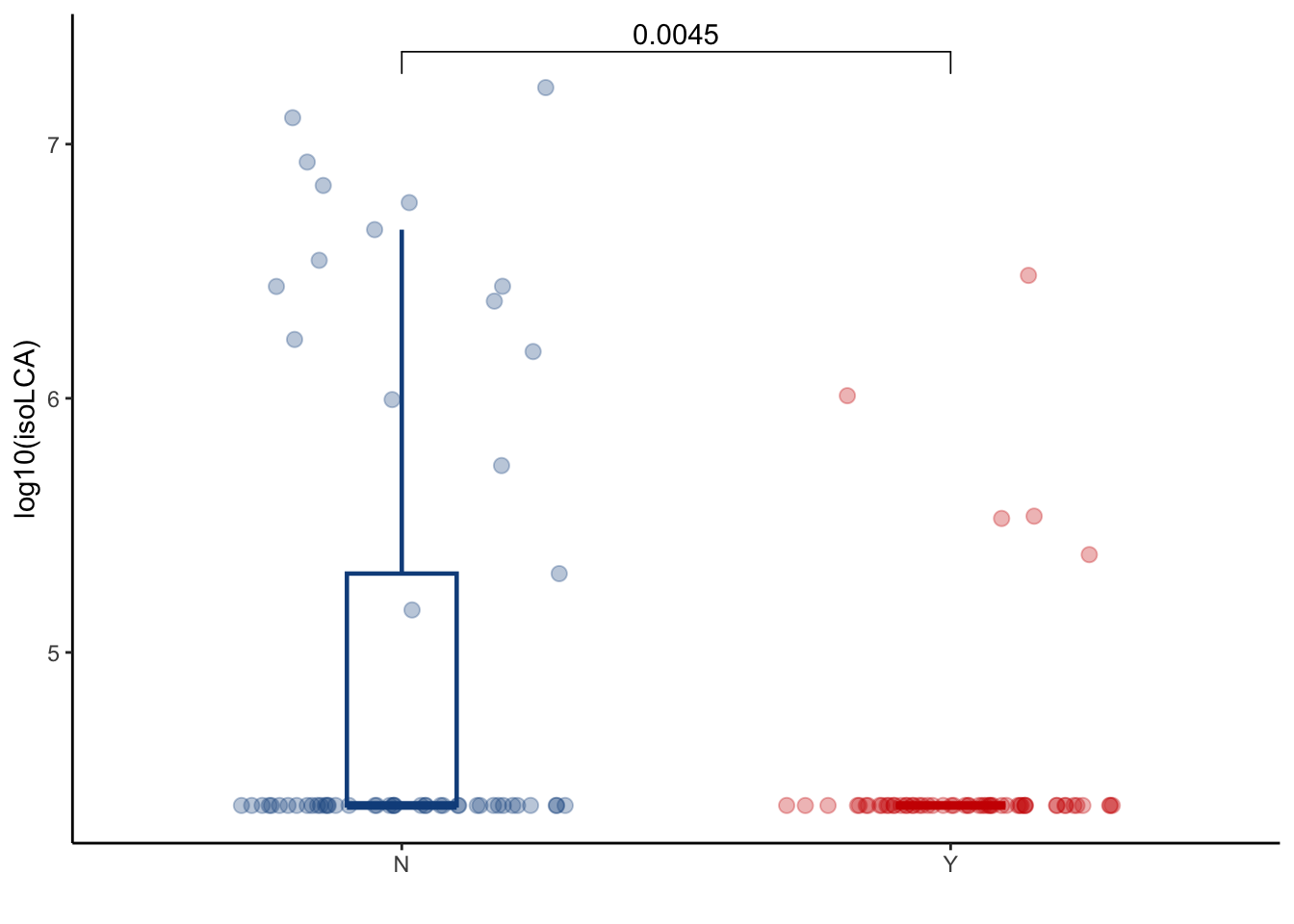
5.3 isoDCA
filtered_combined_table %>%
left_join(cohort_BAS) %>%
filter(later=="Y") %>%
filter(ursodiol=="Y") %>%
ggplot(aes(y=log10(isodeoxycholic_acid+25000), x=GI_GVHD, color=GI_GVHD))+
geom_boxplot(width=0.2, lwd=0.8, outlier.shape = NA) +
geom_jitter(width=0.3, alpha=0.3, size=2.5)+
ylab("log10(isoDCA)")+
xlab("")+
theme_classic()+
stat_compare_means(comparisons=list(c("Y", "N")),
method="wilcox.test",
correct=FALSE)+
scale_color_manual(values=c("dodgerblue4", "red3"))+
theme(legend.position="none")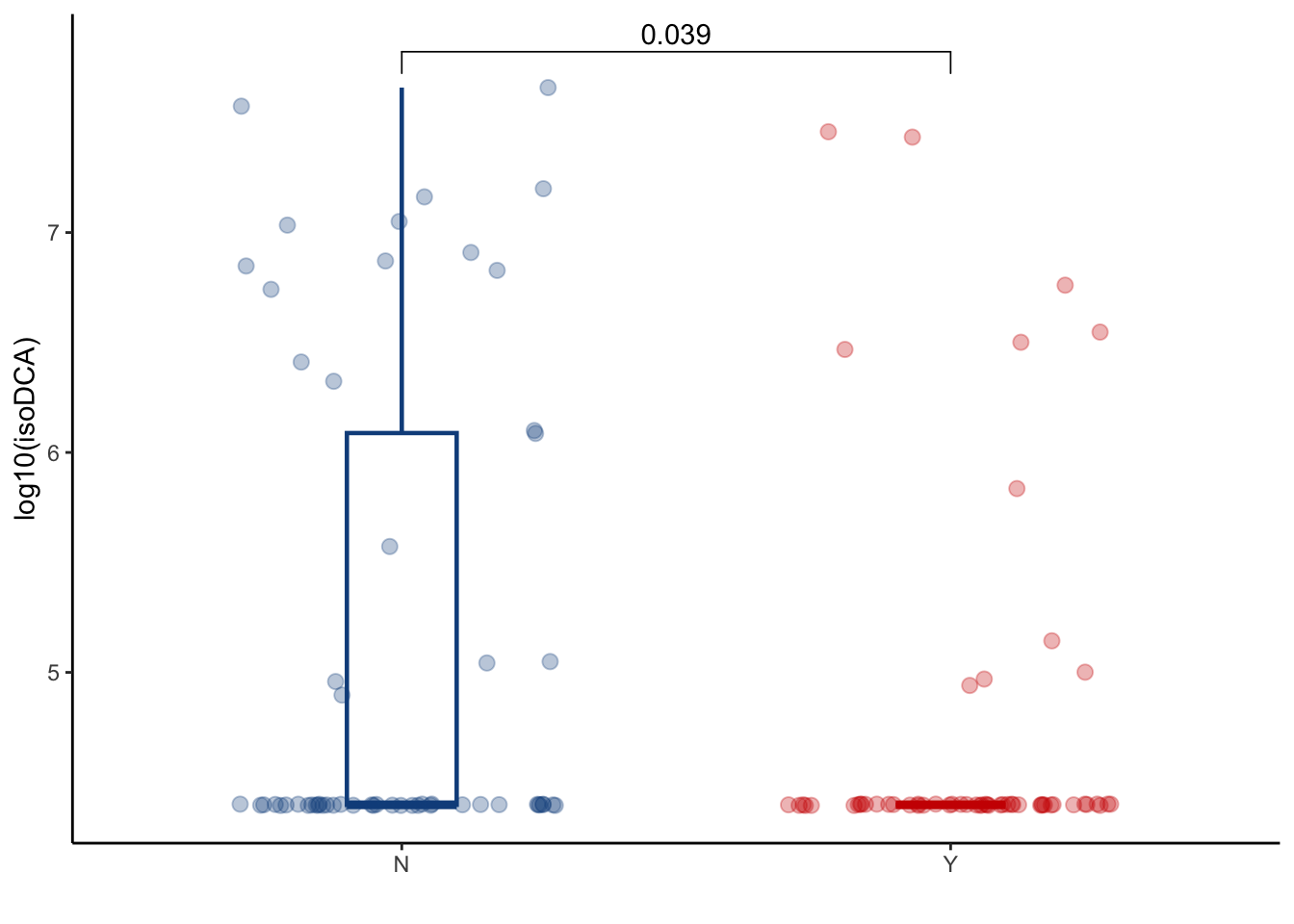
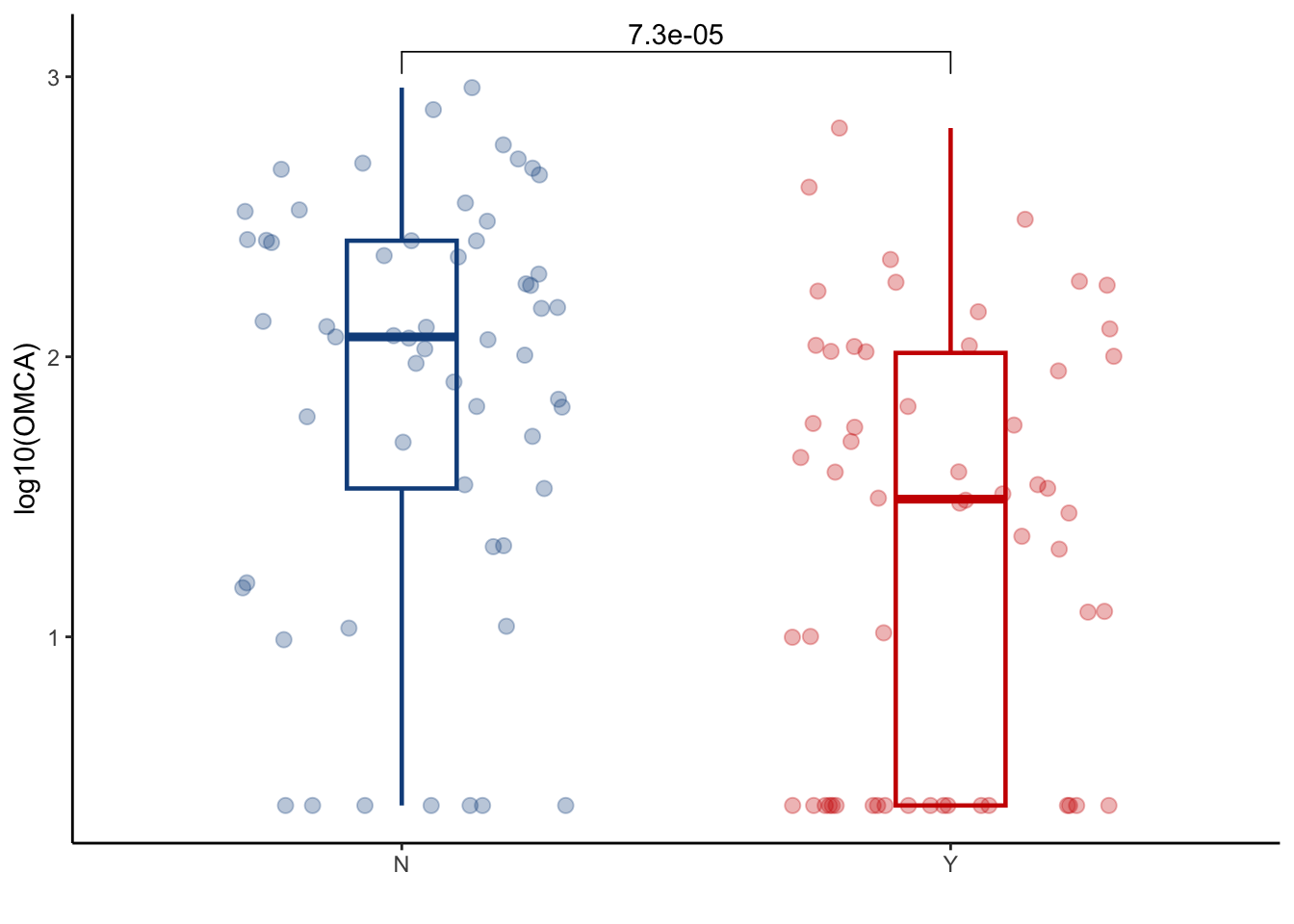
5.4 OMCA
conc_all_filtered %>%
left_join(cohort_BAS) %>%
filter(later=="Y") %>%
filter(ursodiol=="Y") %>%
ggplot(aes(y=log10(omega_muricholic_acid+2.5), x=GI_GVHD, color=GI_GVHD))+
geom_boxplot(width=0.2, lwd=0.8, outlier.shape = NA) +
geom_jitter(width=0.3, alpha=0.3, size=2.5)+
ylab("log10(OMCA)")+
xlab("")+
theme_classic()+
stat_compare_means(comparisons=list(c("Y", "N")),
method="wilcox.test",
correct=FALSE)+
scale_color_manual(values=c("dodgerblue4", "red3"))+
theme(legend.position="none")